Comparing and Combining Results
After you have run each of the Demultiplexing and/or Doublet Detecting softwares you would like, it is helpful to convert them to similar nomenclature and combine the results into a single dataframe. In addition, we have found it helpful to generate summaries of each of the combinations of softwares identified. To help streamline this process, we have provided a script that will easily integrate all the softwares you have run into a single dataframe and can do the following:
Generate a dataframe that has all the software assignments per droplet in the pool
A tab-separated dataframe with the droplet singlet-doublet classification and the individual assignment (for demultiplexing softwares) per droplet
Generate an upset plot that shows the droplet classificaitons by each software and the final classifications
Generate a droplet type summary file
Provides the number of droplets classified for each combination of droplet classifications by each software
Generate demultiplexing individual assignment summary file
Provides the number of droplets classified for each combination of individual assignment droplet classifications by each software
If individuals have not been assigned to each cluster for reference-free demultiplexing softwares, will create a common assignment across all demultiplexing softwares for easy comparison
Combined final droplet assignment from all softwares included
Uses one of four intersectional methods to combine software assignments together into a single combined assignment per barcode
and to generate a summary file for all the software combinations and if you ran demultiplexing softwares, it will also generate a demultiplexing summary file for the individual and cluster assignments from the demultiplexing softwares.
Data
In order to use our script to combine the results from the various demultiplexing and doublet detecting softwares, you need the following:
Required
Output directory (
$OUTDIR)Path to results of each of the softwares you would like to merge into a single dataframe.
You need to provide the path to at least one software result, otherwise, it will not run.
Merging Results with Combine_Results.R
The script has multiple options to provide the paths to each of the software results you would like to run. To see each of the options, simply run:
singularity exec Demuxafy.sif Combine_Results.R -h
Providing the possible parameter options:
usage: /opt/Demultiplexing_Doublet_Detecting_Docs/scripts/Combine_Results.R
[-h] -o OUT [-z DEMUXALOT] [-d DEMUXLET] [-q DROPULATION] ocs/docs/source/Demuxalot Doublet_
[-f FREEMUXLET] [-g FREEMUXLET_ASSIGNMENTS]
[-a FREEMUXLET_CORRELATION_LIMIT] [-s SCSPLIT] [-w SCSPLIT_ASSIGNMENTS]
[-j SCSPLIT_CORRELATION_LIMIT] [-u SOUPORCELL]
[-x SOUPORCELL_ASSIGNMENTS] [-k SOUPORCELL_CORRELATION_LIMIT]
[-v VIREO] [-e DOUBLETDECON] [-t DOUBLETDETECTION] [-i DOUBLETFINDER]
[-n SCDBLFINDER] [-c SCDS] [-r SCRUBLET] [-l SOLO] [-b REF]
[-p PCT_AGREEMENT] [-m METHOD]
optional arguments:
-h, --help show this help message and exit
-o OUT, --out OUT The file where results will be saved
-z DEMUXALOT, --demuxalot DEMUXALOT
Path to demuxalot results. Only use this option if you
want to include the demuxalot results.
-d DEMUXLET, --demuxlet DEMUXLET
Path to demuxlet results. Only use this option if you
want to include the demuxlet results.
-q DROPULATION, --dropulation DROPULATION
Path to dropulation results. Only use this option if
you want to include the dropulation results.
-f FREEMUXLET, --freemuxlet FREEMUXLET
Path to freemuxlet results. Only use this option if
you want to include the freemuxlet results.
-g FREEMUXLET_ASSIGNMENTS, --freemuxlet_assignments FREEMUXLET_ASSIGNMENTS
Path to freemuxlet cluster-to-individual assignments.
Only use this option if have used reference SNP
genotypes to assign individuals to clusters for the
freemuxlet results.
-a FREEMUXLET_CORRELATION_LIMIT, --freemuxlet_correlation_limit FREEMUXLET_CORRELATION_LIMIT
The minimum correlation between the cluster and the
individual SNP genotypes which should be considered as
a valid assignment. If you want no limit, use 0.
Default is 0.7.
-s SCSPLIT, --scSplit SCSPLIT
Path to scSplit results. Only use this option if you
want to include the scSplit results.
-w SCSPLIT_ASSIGNMENTS, --scSplit_assignments SCSPLIT_ASSIGNMENTS
Path to scSplit cluster-to-individual assignments.
Only use this option if you have used reference SNP
genotypes to assign individuals to clusters for the
scSplit results.
-j SCSPLIT_CORRELATION_LIMIT, --scSplit_correlation_limit SCSPLIT_CORRELATION_LIMIT
The minimum correlation between the cluster and the
individual SNP genotypes which should be considered as
a valid assignment. If you want no limit, use 0.
Default is 0.7.
-u SOUPORCELL, --souporcell SOUPORCELL
Path to souporcell results. Only use this option if
you want to include the souporcell results.
-x SOUPORCELL_ASSIGNMENTS, --souporcell_assignments SOUPORCELL_ASSIGNMENTS
Path to souporcell cluster-to-individual assignments.
Only use this option if you have used reference SNP
genotypes to assign individuals to clusters for the
souporcell results.
-k SOUPORCELL_CORRELATION_LIMIT, --souporcell_correlation_limit SOUPORCELL_CORRELATION_LIMIT
The minimum correlation between the cluster and the
individual SNP genotypes which should be considered as
a valid assignment. If you want no limit, use 0.
Default is 0.7.
-v VIREO, --vireo VIREO
Path to vireo results. Only use this option if you
want to include the vireo results.
-e DOUBLETDECON, --DoubletDecon DOUBLETDECON
Path to DoubletDecon results. Only use this option if
you want to include the DoubletDecon results.
-t DOUBLETDETECTION, --DoubletDetection DOUBLETDETECTION
Path to DoubletDetection results. Only use this option
if you want to include the DoubletDetection results.
-i DOUBLETFINDER, --DoubletFinder DOUBLETFINDER
Path to DoubletFinder results. Only use this option if
you want to include the DoubletFinder results.
-n SCDBLFINDER, --scDblFinder SCDBLFINDER
Path to scDblFinder results. Only use this option if
you want to include the scDblFinder results.
-c SCDS, --scds SCDS Path to scds results. Only use this option if you want
to include the scds results.
-r SCRUBLET, --scrublet SCRUBLET
Path to scrublet results. Only use this option if you
want to include the scrublet results.
-l SOLO, --solo SOLO Path to solo results. Only use this option if you want
to include the solo results.
-b REF, --ref REF Which demultiplexing software to use as a reference
for individuals when you do not have assignment key
for all demultiplexing method. Options are 'Demuxlet',
'Freemuxlet', 'scSplit', 'Souporcell' and 'Vireo'. If
blank when assignment keys are missing, default
softwares to use if present are Demuxalot, then Vireo,
then Demuxlet, then Freemuxlet, then Souporcell, then
Dropulation, then scSplit.
-p PCT_AGREEMENT, --pct_agreement PCT_AGREEMENT
The proportion of a cluster that match the 'ref'
assignment to assign that cluster the individual
assignment from the reference. Can be between 0.5 and
1. Default is 0.9.
-m METHOD, --method METHOD
Combination method. Options are 'MajoritySinglet'.
'AtLeastHalfSinglet', 'AnySinglet' or 'AnyDoublet'. We
have found that 'MajoritySinglet' provides the most
accurate results in most situations and therefore
recommend this method. See https://demultiplexing-
doublet-detecting-
docs.readthedocs.io/en/latest/CombineResults.html for
detailed explanation of each intersectional method.
Leave blank if you just want all the softwares to be
merged into a single dataframe.
Combination Methods - Additional Information
There are four options for making combined droplet type (singlet or doublet) and individual assignment from the softwares used:
MajoritySinglet
If more than half of the softwares identify a droplet as a singlet, it is classified as a singlet.
If more than half the demultiplexing softwares identify the same indivdual, that assignment is used for the droplet.
We have found
AtLeastHalfSinglet
If at least half of the softwares identify a droplet as a singlet, it is classified as a singlet.
If at least half the demultiplexing softwares identify the same indivdual, that assignment is used for the droplet.
AnySinglet
If this droplet is identified as a singlet by any software, the droplet is classified as a singlet.
In other words, a doublet is only called if all softwares identified that droplet as a doublet.
AnyDoublet
A droplet is classified as a singlet only if all softwares identify it as a singlet.
In other words, a doublet is called if any software identifies that droplet as a doublet.
An example command that combines Demuxlet results, Souporcell results, Solo results and Scds results would look like this: There are a two different options for using this script: 1. Combining the results and calling the droplet type through the combination of the softwares. 1. Combining the results without any joint droplet calling. You might choose this if you just want to see how the different softwares perform on your data before deciding which to move forward with for final joint droplet calling.
First, let’s assign the variables that will be used to execute each step.
Example Variable Settings
Below is an example of the variables that we can set up to be used in the command below. These are files provided as a test dataset available in the Data Preparation Documentation Please replace paths with the full path to data on your system.
OUTDIR=/path/to/output/combined DEMUXLET_OUTDIR=/path/to/output/demuxlet SOUPORCELL_OUTDIR=/path/to/output/souporcell SOLO_OUTDIR=/path/to/output/solo SCDS_OUTDIR=/path/to/output/scds
The first option is to select a method to make joint calls on the individual assignment and singlet-doublet droplet types using the softwares included.
singularity exec Demuxafy.sif Combine_Results.R \
-o $OUTDIR/combined_results.tsv \
--demuxlet $DEMUXLET_OUTDIR \
--souporcell $SOUPORCELL_OUTDIR \
--solo $SOLO_OUTDIR \
--scds $SCDS_OUTDIR \
--method "MajoritySinglet" ## there are other methods that can also be used, please see the help message above for the other options
The other option is to just combine the results together without instersectional joint calls on the assignment and droplet type for each droplet.
singularity exec Demuxafy.sif Combine_Results.R \
-o $OUTDIR/combined_results.tsv \
--demuxlet $DEMUXLET_OUTDIR \
--souporcell $SOUPORCELL_OUTDIR \
--solo $SOLO_OUTDIR \
--scds $SCDS_OUTDIR
Note
The path to the directories will work if the file names are the expected file names based on the example tutorials. However, if you used a different file naming convention or changed the names, you can also provide the full path to the exact file for each software.
Results and Interpretation
After running the Combine_Results.R script, you should have two, three or four files depending on if you used demultiplexing softwares and if you used joint droplet calling.
Here, we show the results for the above example that also provides combined calls with the “MajoritySinglet” calls.
/path/to/output/combined
├── combined_results_demultiplexing_summary.tsv
├── combined_resultsSinglets_upset.pdf
├── combined_results_summary.tsv
├── combined_results.tsv
└── combined_results_w_combined_assignments.tsv
Note
You will only have the
combined_results_demultiplexing_summary.tsvfile if you included demultiplexing softwares.And you will only have the
combined_results_w_combined_assignments.tsvfile if you ran it with--method
Here’s a deeper look at the contents of each of these results:
combined_resultsSinglets_upset.pdf
This is an upset figure of the droplets which are colored by their finall individual or doublet classification.
A filled circle indicates the that those droplets are classified as singlets by that method while empty circles indicate a doublet classification by that software
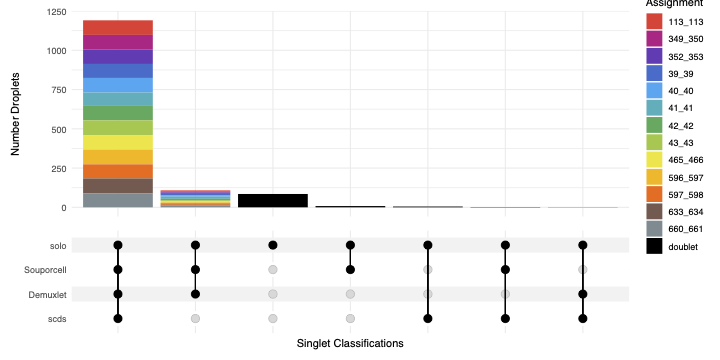
combined_results.tsv
Has the selected results combined; only including key columns.
Barcode
Demuxlet_DropletType
Demuxlet_Individual_Assignment
Souporcell_Individual_Assignment
Souporcell_Cluster
Souporcell_DropletType
scds_score
scds_DropletType
solo_DropletType
solo_DropletScore
AAACCTGAGATAGCAT-1
singlet
41_41
41_41
6
singlet
0.116344358493288
singlet
singlet
-8.442187
AAACCTGAGCAGCGTA-1
singlet
465_466
465_466
11
singlet
0.539856378453988
singlet
singlet
-2.8096201
AAACCTGAGCGATGAC-1
singlet
113_113
113_113
5
singlet
0.0237184380134577
singlet
singlet
-2.8949203
AAACCTGAGCGTAGTG-1
singlet
349_350
349_350
3
singlet
0.163695865366576
singlet
singlet
-5.928284
AAACCTGAGGAGTTTA-1
singlet
632_633
632_633
7
singlet
0.11591462421927
singlet
doublet
0.2749935
AAACCTGAGGCTCATT-1
singlet
39_39
39_39
12
singlet
0.0479944175570073
singlet
singlet
-5.2726507
AAACCTGAGGGCACTA-1
singlet
465_466
465_466
11
singlet
0.374426050641161
singlet
singlet
-0.65760195
AAACCTGAGTAATCCC-1
singlet
660_661
660_661
4
singlet
0.247842972104563
singlet
singlet
-3.5948637
AAACCTGAGTAGCCGA-1
doublet
doublet
unassigned
unassigned
unassigned
0.342998285281922
singlet
singlet
-0.50507957
…
…
…
…
…
…
…
…
…
…
combined_results_summary.tsv
The number of each of the combinations of the software cell type classifications
Demuxlet_DropletType
Souporcell_DropletType
scds_DropletType
solo_DropletType
N
singlet
singlet
singlet
singlet
16193
doublet
doublet
doublet
doublet
1714
singlet
singlet
singlet
doublet
947
doublet
doublet
singlet
singlet
468
singlet
singlet
doublet
singlet
392
singlet
singlet
doublet
doublet
345
doublet
doublet
singlet
doublet
335
doublet
singlet
singlet
singlet
171
doublet
doublet
doublet
singlet
169
doublet
singlet
doublet
doublet
114
doublet
singlet
singlet
doublet
44
doublet
singlet
doublet
singlet
18
singlet
doublet
singlet
singlet
17
singlet
unassigned
singlet
singlet
13
doublet
unassigned
singlet
singlet
11
singlet
doublet
doublet
doublet
9
singlet
doublet
singlet
doublet
6
singlet
doublet
doublet
singlet
5
doublet
unassigned
singlet
doublet
4
doublet
unassigned
doublet
doublet
3
doublet
unassigned
doublet
singlet
2
unassigned
unassigned
singlet
singlet
2
combined_results_demultiplexing_summary.tsv
Summary of the number of each of the combination of classifications by demultiplexing software:
Demuxlet_Individual_Assignment
Souporcell_Individual_Assignment
N
doublet
doublet
2706
352_353
352_353
1603
43_43
43_43
1547
597_598
597_598
1510
349_350
349_350
1450
42_42
42_42
1417
660_661
660_661
1358
113_113
113_113
1333
39_39
39_39
1289
…
…
…
combined_results_w_combined_assignments.tsv
Dataframe combining all the software results together + combined assignment based on selected method:
Barcode
Demuxlet_DropletType
Demuxlet_Individual_Assignment
Souporcell_Cluster
Souporcell_Individual_Assignment
Souporcell_DropletType
scds_score
scds_DropletType
solo_DropletType
solo_DropletScore
MajoritySinglet_DropletType
MajoritySinglet_Individual_Assignment
AAACCTGAGATAGCAT-1
singlet
41_41
6
41_41
singlet
0.116344358493288
singlet
singlet
-8.442187
singlet
41_41
AAACCTGAGCAGCGTA-1
singlet
465_466
11
465_466
singlet
0.539856378453988
singlet
singlet
-2.8096201
singlet
465_466
AAACCTGAGCGATGAC-1
singlet
113_113
5
113_113
singlet
0.0237184380134577
singlet
singlet
-2.8949203
singlet
113_113
AAACCTGAGCGTAGTG-1
singlet
349_350
3
349_350
singlet
0.163695865366576
singlet
singlet
-5.928284
singlet
349_350
AAACCTGAGGAGTTTA-1
singlet
632_633
7
632_633
singlet
0.11591462421927
singlet
doublet
0.2749935
singlet
632_633
AAACCTGAGGCTCATT-1
singlet
39_39
12
39_39
singlet
0.0479944175570073
singlet
singlet
-5.2726507
singlet
39_39
AAACCTGAGGGCACTA-1
singlet
465_466
11
465_466
singlet
0.374426050641161
singlet
singlet
-0.65760195
singlet
465_466
AAACCTGAGTAATCCC-1
singlet
660_661
4
660_661
singlet
0.247842972104563
singlet
singlet
-3.5948637
singlet
660_661
AAACCTGAGTAGCCGA-1
doublet
doublet
unassigned
doublet
doublet
0.342998285281922
singlet
singlet
-0.50507957
doublet
doublet
…
…
…
…
…
…
…
…
…
…
…
…
Citation
If you used the Demuxafy platform for analysis, please reference our preprint.